Department of Infectious Diseases and Immunology
National Hospital Organization Nagoya Medical Center partly functions as an “AIDS Care Core Hospital” in Tokai district (Gifu, Shizuoka, Aichi, and Mie Prefectures) to provide the state-of-the-art medical services for HIV/AIDS. The Clinical Research Center resides in the hospital, devoting medical and clinical research. Our research group, department of “Infectious Diseases and Immunology”, investigates both on basic and clinical sciences for HIV/AIDS and the related pathogen-infectious diseases, aiming at rapid translations of scientific observations and laboratory discoveries into new approaches for diagnosing, treating, and preventing HIV infection and progress to AIDS.
In Japan, more than 1,400 people per year are newly diagnosed with HIV today, and unfortunately, the incidence of new HIV infection is still increasing year by year, particularly around Nagoya area. Recent chemotherapies using combinations of more than three antiretrovirals (combination antiretroviral therapy: cART) have improved the prognosis of the HIV seropositive people. However, since cART should be continued for life-long period, we must consider the presumable problem facing the appearance and development of resistance in the virus. To avoid such problem, it is quite requisite to understand how the virus develops resistance to drugs, to survey how such drug-resistant viruses spread in Japan epidemiologically, and to develop more effective strategies for treating HIV infection. Therefore, our research team devotes all the efforts to investigate the fundamental studies by setting up the following five specific projects; 1) Epidemiological survey of drug-resistant HIV in Japan, collaborating with prefectural and municipal public health institutes, National Institutes of Infectious Diseases, and Universities. 2) Analysis of molecular/structure-biological mechanism for drug-resistant viruses, 3) Development of anti-HIV compounds having novel targets, 4) Epidemiological and genetical investigations of infectious pathogens related to HIV infection, such as Hepatitis B, C viruses. 5) Analysis of molecular mechanism on anti-HIV factors derived from the host, such as APOBEC3s, and the application/development for new anti-HIV therapies.
In addition to HIV/AIDS research, our department has joined in a medical education program as an “affiliated laboratory” of the Nagoya University Graduate School of Medicine since 2009 (AIDS Research laboratory)
Research Topic 1: Epidemiological Investigation of Drug-resistant HIV in Japan
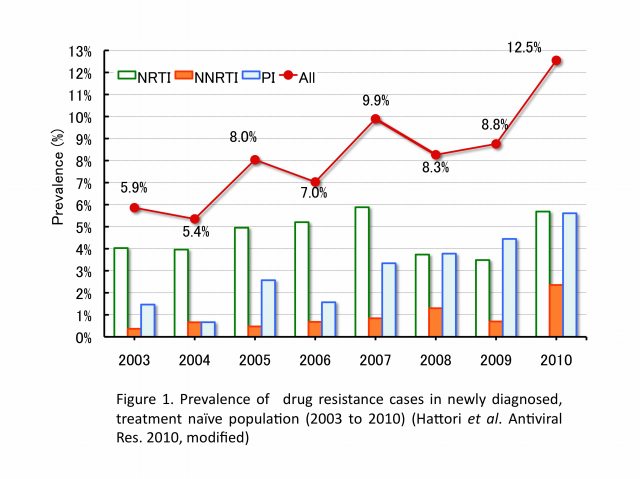
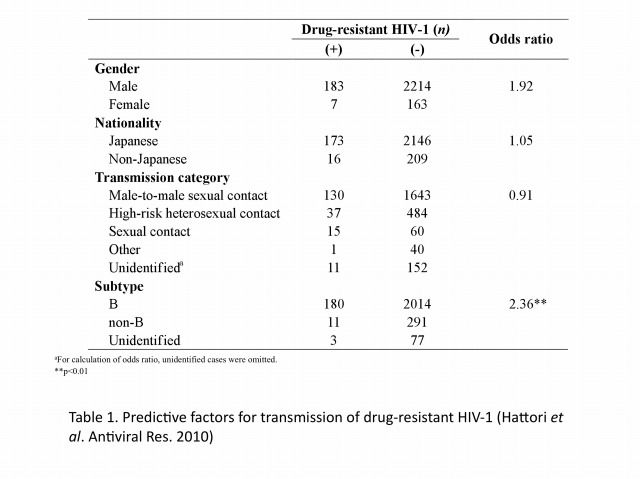
The emergence and transmission of drug-resistant HIV-1 compromise anti-retroviral treatment for HIV-1. Thus, testing for drug resistance is recommended when diagnosed as HIV seropositive and before starting cART. Our group has been conducting an epidemiological survey enrolling newly diagnosed patients since 2003 in collaboration with our nationwide surveillance network. In the initial 6-year study period, the prevalence of drug-resistant HIV-1 among 2573 patients, consisting mainly of Japanese men in their late-30s and infected through male-to-male sexual contacts, followed an increasing trend from 5.9% (16/273) in 2003 to 8.3% (50/605) in 2010 (Figure 1). Nucleoside reverse transcriptase inhibitor-associated mutations predominated in each year, with T215 revertants being the most abundant. The predictive factor for drug-resistant HIV-1 transmission was subtype B and those for recent HIV-1 infection were male gender, MSM behavior, Japanese nationality, and subtype B (Table 1). Continued activities are needed to raise awareness of the risks of HIV-1 infection and complications of drug-resistant strains. Continued surveillance is also needed to understand trends in the HIV-1 epidemic.
Research Topic 2: Analysis of Molecular Mechanism on Drug-resistant HIV
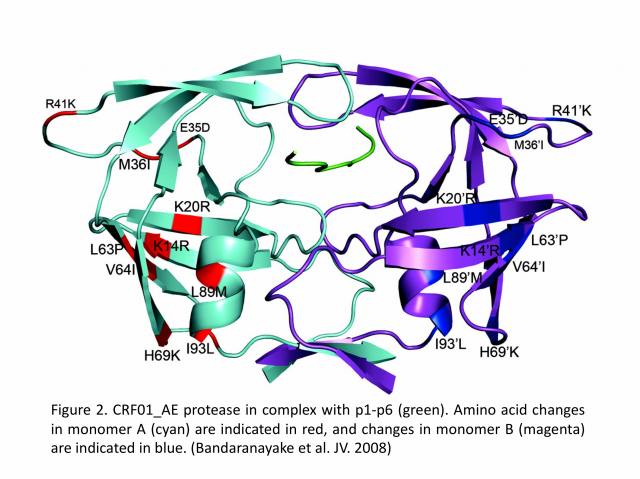
Recently, there has been a greater interest in what is the effect of amino acid variability between HIV-1 clades on structure and the emergence of resistance mutations in HIV-1. In collaboration with Dr. Celia A. Schiffer’s group (University of Massachusetts Medical School, USA), we determined the first crystal structure of the HIV-1 CRF01_AE protease in complex with the p1-p6 substrate (Figure 2). The results show that structural rearrangements in CRF01_AE protease appear by hydrogen bonding between the flap hinge and the protease core regions, which is quite significant compared to the subtype-B protease structure.
Research Topic 3: Development of Novel Anti-HIV Drugs and Therapies
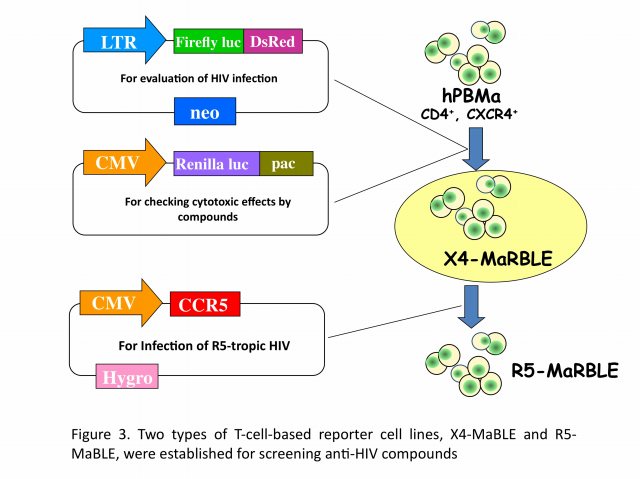
We have developed two new T-cell-based reporter cell lines, R5-MaBLE and X4-MaBLE, to enable to perform high-throughput screening for potential anti-HIV compounds (Figure 3). One cell line naturally expresses CD4 and CXCR4, making it susceptible to X4-tropic viruses, and the other cell line, in which a CCR5 expression vector was introduced, is susceptible to both X4- and R5-tropic viruses. Reporter cells were constructed by transfecting the human T-cell line HPB-Ma, which demonstrates high susceptibility to HIV-1, with genomes expressing two different luciferase reporters, HIV-1 long terminal repeat-driven firefly luciferase and cytomegalovirus promoter-driven renilla luciferase. Upon HIV infection, the cells expressed firefly luciferase at levels that were highly correlated with the production of the capsid antigen p24. The cells also constitutively expressed renilla luciferase, which was used to monitor cell numbers and viability. The reliability of the cell lines for two in vitro applications, drug resistance phenotyping and drug screening, was confirmed. As HIV-1 efficiently replicated in these cells, they could be used for multiple-round replication assays as an alternative method to a single-cycle replication protocol. By using our unique assay systems, we are currently exploring new anti-HIV chemicals for future.
Research Topic 4: Epidemiological and Genetical Investigations of HBV co-infected with HIV-1
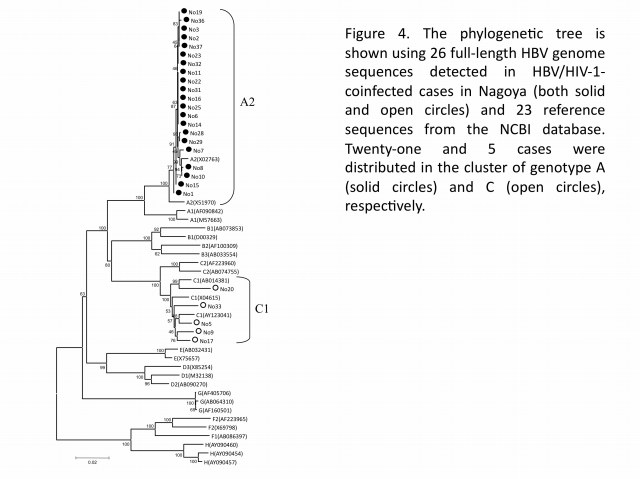
In hepatitis B virus (HBV) infection, mother-to-child transmission (MTCT) and blood transfusion has been regarded as the major routes in Japan. However, the incidence of HBV tarnsmission through sexual contacts appears to be increasing, especially among HIV-1-seropositive patients. To better understand the molecular epidemiology of HBV co-infected with HIV-1, our group analyzed both HBV genotypes and HIV-1 subtypes among HBV/HIV-1-coinfected cases at Nagoya Medical Center from 2003 to 2007. Among 394 HIV-1-positive cases newly diagnosed during the study period, it turned out 31 (7.9%) positive for the hepatitis B surface antigen, which were Japanese men having sex with men (MSM). Analysis of the HBV genome sequences (26 cases) revealed that 21 (80.7%) and 5 (19.3%) cases were determined as genotypes A and C (Figure 4), respectively, indicating that HBV genotype A was dominant in HIV-1-seropositive patients. This finding alerts clinicians to an alternative outbreak of HBV genotype A in the HIV-1-infected MSM population, and to a shift of the genotype from C to A in Japan. The narrow genetic diversity in genotype A suggests that the particular genotype has been recently introduced into the MSM population. Moreover, we found a lamivudine-resistant mutation in 1-naive case, showing a potential risk of drug-resistant HBV transmission. These studies suggest that infection of HBV genotype A could have a higher risk than the other for individuals to become HBV carriers, and that prevention programs are urgently needed for the targeted population in Japan.
Research Topic 5: Analysis of Molecular Mechanism for Anti-HIV and Retroviral Host Factors
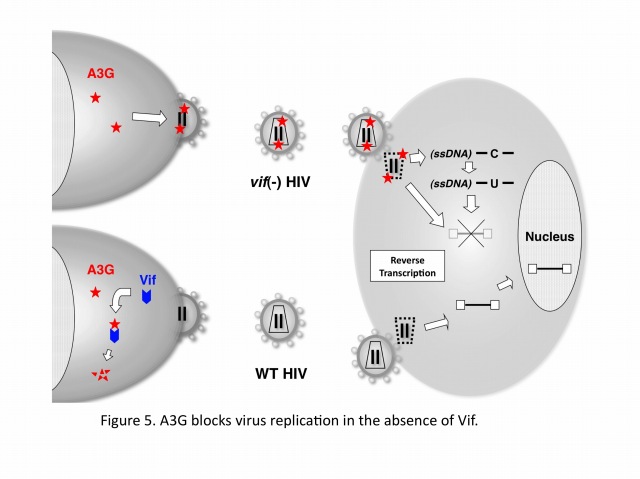
Human cells developed the defense systems against retrovirus infections during the evolutions. These systems include retroviral restrictions by DNA cytidine deaminases of APOBEC3 family (A, B, C, DE, F, G, and H), which are potent factors to block the viral replication by blocking reverse transcription and/or integration and by hypermutating viral cDNA. In case of HIV-1, the viral protein, Vif abrogates the APOBEC3F/G function through specific machinery of ubiquitination and proteasomal degradation. Without Vif, APOBEC3F/G are incorporated into virus particles and block reverse transcription and/or integration in a newly infected cell. Recent advances in our understanding about biochemical and structure-biological characteristics of the enzymes provide new insights to reveal more detailed molecular mechanisms for anti-retroviral activity by APOBEC3 family.
APOBEC3G (A3G) is a host cytidine deaminase that serves as a cellular defense protein and a potent inhibitor of HIV-1 replication. Abrogation of A3G function involves the HIV-1 Vif protein, a specific substrate receptor for ubiquitination that binds A3G. More specifically, Vif facilitates ubiquitination of A3G by forming an E3 complex consisting of Cullin5-ElonginB/C-Rbx2-E2, which targets A3G for rapid proteasomal degradation. Yet, despite our knowledge that these proteins interact with each other in the complex, the details of how the complex dictates A3G ubiquitination and subsequent degradation have remained unclear.
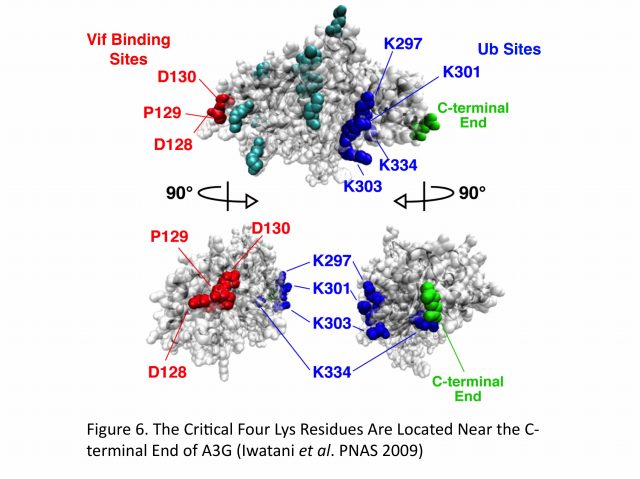
Extensive mutagenesis, guided by a structural model of full-length human A3G, has allowed us for the first time to identify four Lys residues (Lys297, 301, 303, and 334) that are required for HIV-1 Vif-dependent ubiquitination and degradation. Remarkably, substitution of Arg for Lys at these residues (the mutant is termed super A3G [S-A3G]) confers Vif resistance and restores A3G’s antiviral activity even in the presence of Vif. In addition, we found that the interaction between WT A3G and S-A3G with Vif is very similar and that S-A3G is incorporated into WT and vif-deficient viral particles to the same extent. Interestingly, the Vif sensitivity of S-A3G was changed by addition of the C-terminal tags, indicating that Lys residues in the C-terminal tag can serve as ubiquitination sites. In our A3G structure model, the critical four residues cluster at the C-terminus, which is opposite to the known N-terminal A3G-Vif interaction region containing the 128DPD130 motif (Figure 5). Moreover, HIV-2 and SIVmac Vif proteins target the same Lys residues in A3G as HIV-1 Vif. Collectively, these findings suggest that spatial constraints imposed by the E3 complex and especially the rigid scaffold Cullin5 protein may be an important determinant for Vif-dependent A3G ubiquitination/degradation (Figure 6).